The Krishnamurthy Lab
The Story Behind the Depsipeptide Project in the CCE

The depsipeptide project is a joint and collaborative effort between many groups within the Center for Chemical Evolution (CCE).
In response to questions from Juila Eckhoff, associate editor at Nature Chemistry Reviews (in the context of the PNAS paper http://dx.doi.org/10.1073/pnas.1711631114 (2017) Jay Forsythe, Ram Krishnamurthy and Facundo Fernandez put together the following:
Can you give me some background on this project?
The proto-peptide project (or “depsipeptide project” as we call it internally in our Center) is one of the central approaches towards emergence of proto-biopolymers taken by the Center for Chemical Evolution since 2014 (see below for the prior work that led to this project). In this PNAS paper we wanted to have a more realistic approach in terms of complexity to understand how more organized systems would arise and evolve when starting from mixtures of a spectrum of prebiotic building blocks. The ester-amide exchange chemistry that we have been investigating as a means to generate proto-peptides was in a unique position to generate such a complex mixture that better resembles the “primordial soup”. It also lent itself nicely to the mass spectrometry analytical techniques that are in place for proteomics, allowing for real time evolution and monitoring of sequence-space complexity. However, we soon realized that we needed to fill some gaps in the existing analytical approaches and generate our own pipeline and algorithms for analyzing the complexity generated by the depsipeptide system, which far outweighs that of biological peptides by orders of magnitude.
Why did you decide to do this project? Also, what excites you most about this area of research?
We realized that the field of prebiotic chemistry had not benefitted from omics technologies that are commonplace in systems biology, and that we could learn a great deal from the insights that we could obtain from looking at much more complex systems than those looked at traditionally in the origins of life field. What is really exciting is that the toolbox is now available to tackle questions in chemical evolution that were previously unanswerable. I feel we are very close to understanding how informational and functional (bio)polymers emerged from the primordial soup, shedding some light onto the bigger question on how life emerged on Earth.
What prior work led to these discoveries?
The “depsipeptide project” was started in 2014 and was based on an idea proposed by Ram Krishnamurthy at The Scripps Research Institute during one of our weekly CCE meetings while listening to the presentation by Irena Mamajanov, a post-doctoral colleague in the lab of Nick Hud. The Hud lab was working on the dry-wet cycling elongation of malic acid, based on the hypothesis that the polyesters from alpha-hydroxy acids could have acted as prebiotic catalysts before the arrival of polypeptides, an idea that has its foundation in the work of Alex Rich at MIT (A. Rich in Chemical Evolution and the Origin of Life (Eds.: R. Buvet, C. Ponnamperuma), North-Holland, Amsterdam, 1971, pp. 180 – 196). The Hud group together with my group and Martha Grover’s had shown that malic acid polymerizes readily under potential prebiotic under mild heat of the dry-wet cycling process to give rise to polyesters (Macromolecules 2014, 47, 1334 – 1343.), a process that does not produce oligopeptides when starting from only amino acids. Ram Krishnamurthy proposed that starting with a mixture that included amino acids and alpha-hydroxy acids (which is a prebiotic reality as documented in many of the meteorites and the Miller-Urey spark discharge experiments, see Rapid Commun Mass Spectrom., 2016, 30(18), 2043-51 ), the wet-dry cycling process would lead to depsipeptides, which on continued cycling would gradually increase in their peptide content based on an exchange of the ester bond with an amide bond.
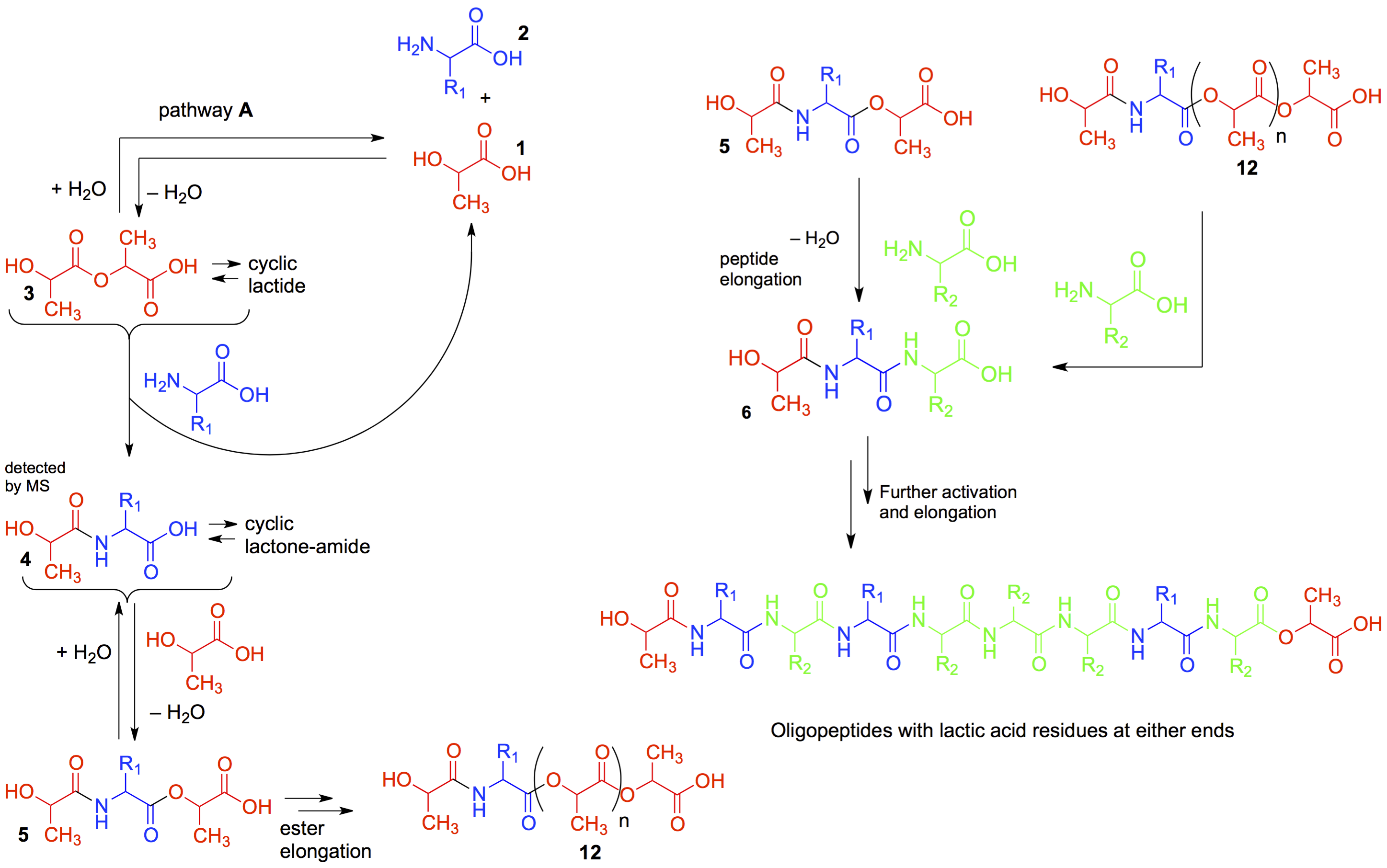
This expectation is based on the simple fact that ester bonds (which are formed easily since the thermodynamic barrier to an ester bond is lower), would be reactive towards nucleophiles such as amino acids. The attack of the NH2 group of the amino acid on the ester bond would produce a peptide bond. The free carboxylic acid group of this newly formed (depsi)peptide can react further with another alpha-hydroxy acid to make another ester bond. This new ester bond would get attacked again by another amino acid. Repeating this process in cyclic steps would lead to longer oligo(depsi)peptides enriched in peptide bonds. Moreover, peptides bonds once formed are thermodynamically more stable ( compared to ester bonds) and, thus, would be expected to accumulate when compared to the more hydrolytically labile ester bonds. Once the project started with Sheng-Sheng Yu (from Martha Grover’s lab) the project took off. It was soon realized that to analyze and sequence the complex mixtures of depsipeptides that were being formed, sophisticated analytical techniques would be needed, and Jay Forsythe (from Facundo's lab) got involved and took a leadership role in formulating experiments and the mass spectrometry driven analysis. From then on it rapidly progressed to a publication (Angew. Chem. Int . Ed. 2015, 54, 9871-9875), which demonstrated that the ester-mediated amide bond formation driven by wet-dry cycles could be a possible path to polypeptides on prebiotic earth, without the need for any extreme temperatures or other external chemical activating agents to drive the condensation of amino acids.
Can you explain some of the methodology?
We used a hybrid multi-dimensional separation approach. Parts of it was borrowed directly from the proteomics community. Traditionally, this community uses liquid chromatography coupled to tandem mass spectrometry to sequence peptides obtained from enzymatic digestion of proteins, followed by database searching. These databases, however, do not exist for the types of peptides we were interested in (depsipeptides), so we had to come up with our custom databases and search algorithm. We also added a dimension of ion mobility separation to increase the number of species we could separate, and look at their emerging folding by measuring their gas-phase collision cross sections. Overall, it was an analytical tour de force!
And why did your choice fall on G, A and L as a set of amino acids for this approach?
The choices were mainly dictated by the prebiotic availability of the starting building blocks as seen in the meteorites and in many of the prebiotic simulations (such as the Miller-Urey spark discharge) that produced amino acids and alpha-hydroxy acids. We felt that thee amino acids and one hydroxy-acid mixtures were all we wanted to tackle initially. And we detected hundreds of depsipeptides from this simple starting materials. We are now looking into mixtures that are formed with many more building blocks (11 amino acids and 4 hydroxy acids mixed together).
Further, I am curious about: What is next? What are the implications for future research and applications?
We are currently investigating the properties of these depsipeptides in their abilities to (a) form structures, (b) to act as catalysts and (c) to associate with other proto-biomolecules to enable self-assembly. A tantalizing possibility would be that there arises a depsi(peptide) catalyst capable of making peptide bonds. This would be a game changer as the system can become free from the environment and could become a self-driven process from within.

